-
- Downloads
real comparison code
Showing
- pybasicbayes/distributions/comparison_results 0 additions, 0 deletionspybasicbayes/distributions/comparison_results
- pybasicbayes/distributions/dynglm_optimisation_test.py 49 additions, 0 deletionspybasicbayes/distributions/dynglm_optimisation_test.py
- pybasicbayes/distributions/test_data 0 additions, 0 deletionspybasicbayes/distributions/test_data
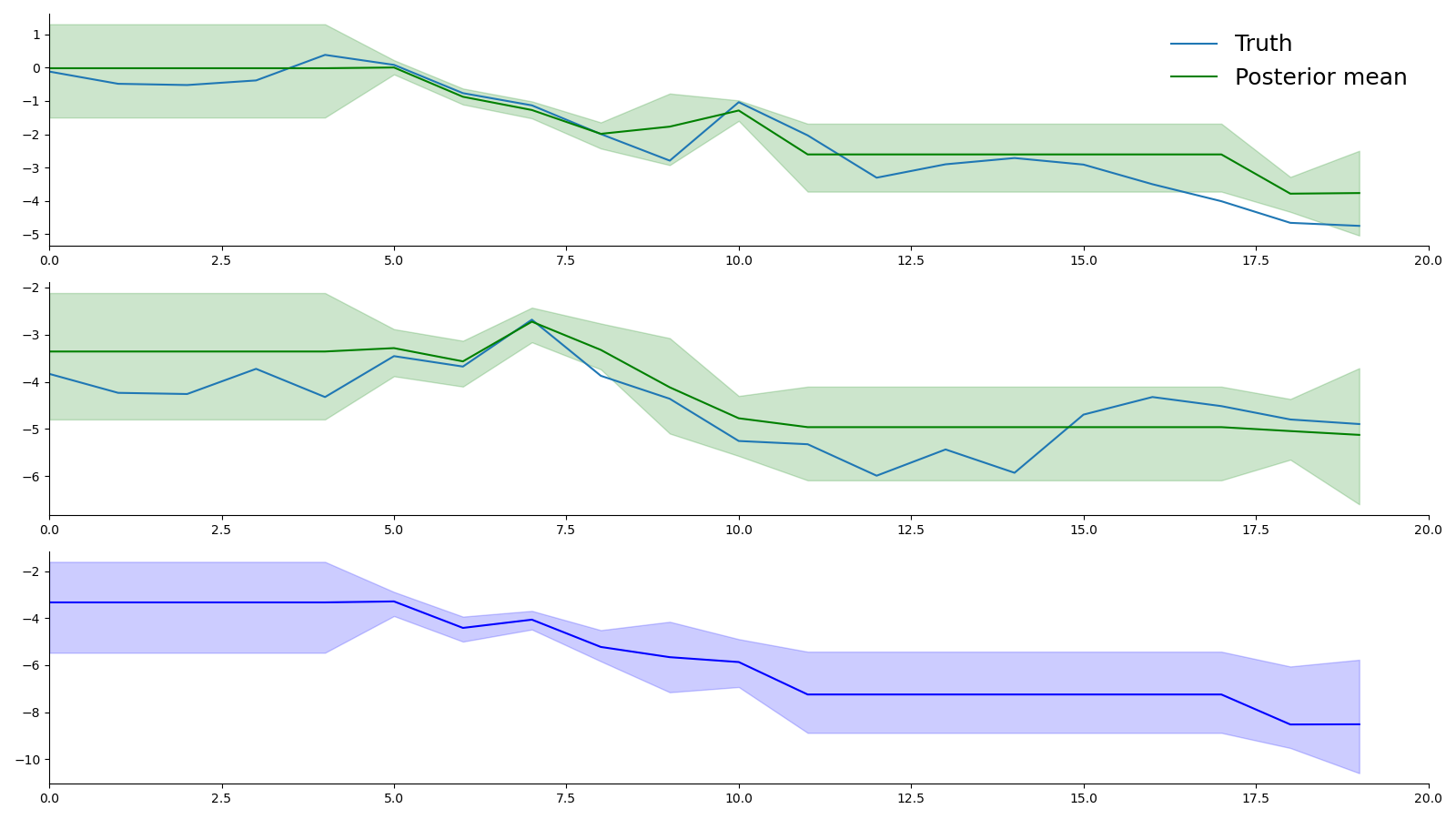
- pybasicbayes/distributions/timepoint test.png 0 additions, 0 deletionspybasicbayes/distributions/timepoint test.png
- pybasicbayes/distributions/truth 0 additions, 0 deletionspybasicbayes/distributions/truth
File added
pybasicbayes/distributions/test_data
0 → 100644
File added
106 KiB
pybasicbayes/distributions/truth
0 → 100644
File added