diff --git a/README.md b/README.md
index 4c5e6f3286dff3626a0427a60bf91f814ded9c0a..075103bd6ca907f7bbb907650c167fdb66610df3 100644
--- a/README.md
+++ b/README.md
@@ -71,7 +71,7 @@ data = np.loadtxt('data.txt')
plt.plot(data[:,0],data[:,1],'kx')
```
-
+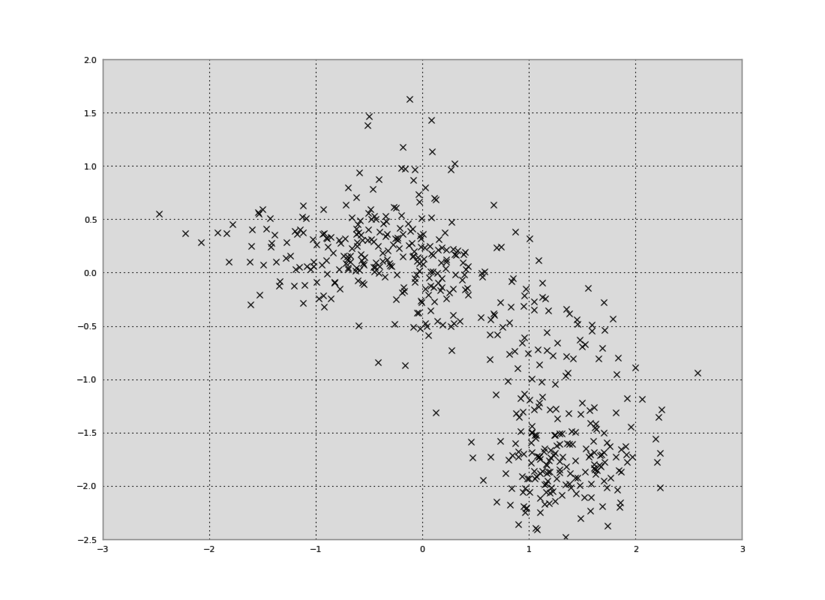
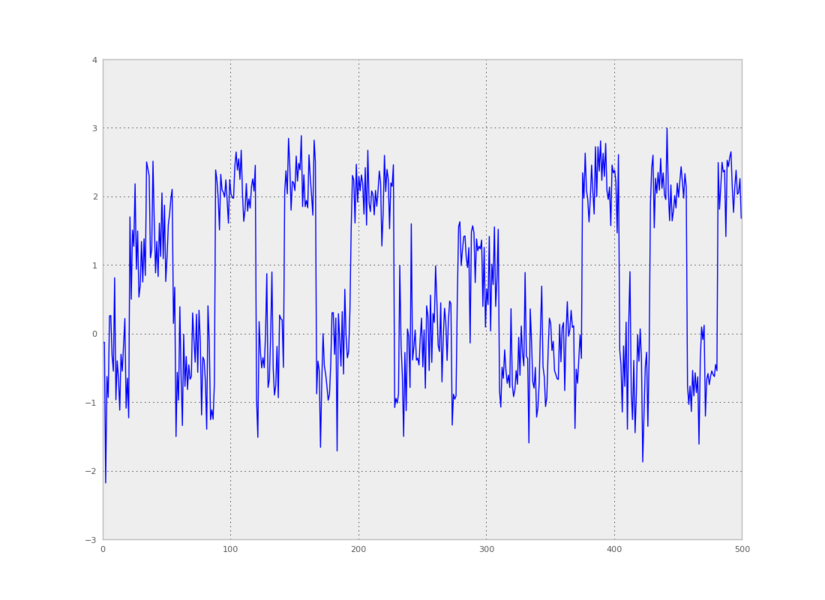
We can also make a plot of time versus the first principal component:
@@ -80,7 +80,7 @@ from pyhsmm.util.plot import pca_project_data
plt.plot(pca_project_data(data,1))
```
-
+
To learn an HSMM, we'll use `pyhsmm` to create a `WeakLimitHDPHSMM` instance
using some reasonable hyperparameters. We'll ask this model to infer the number
@@ -156,11 +156,11 @@ for idx, model in enumerate(models):
plt.savefig('iter_%.3d.png' % (10*(idx+1)))
```
-
+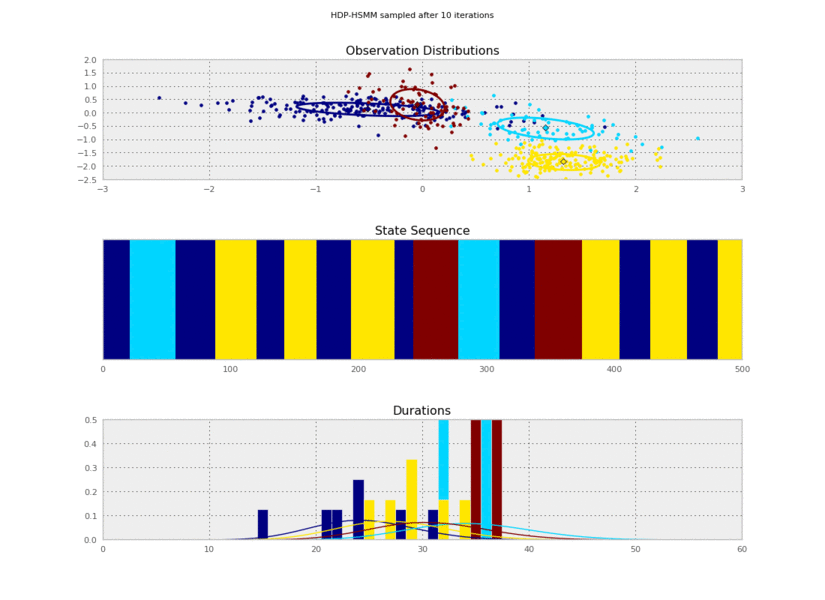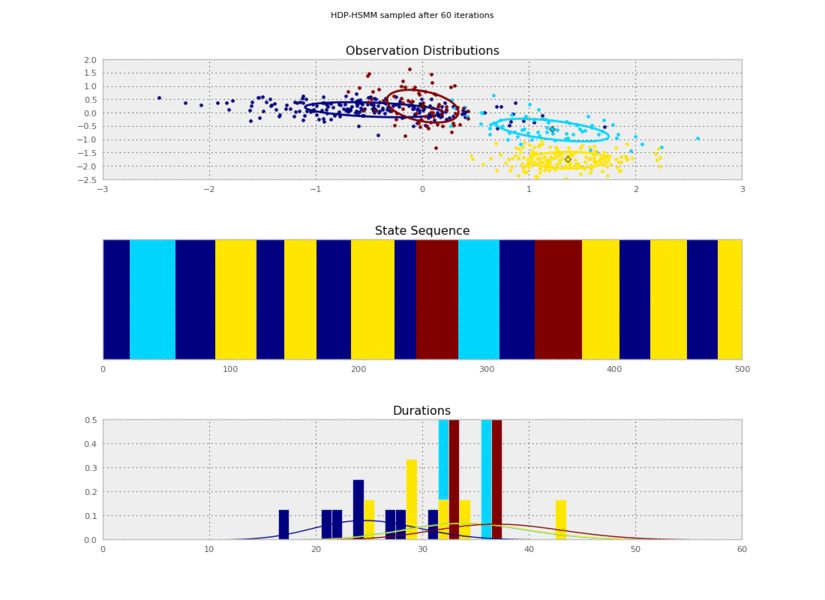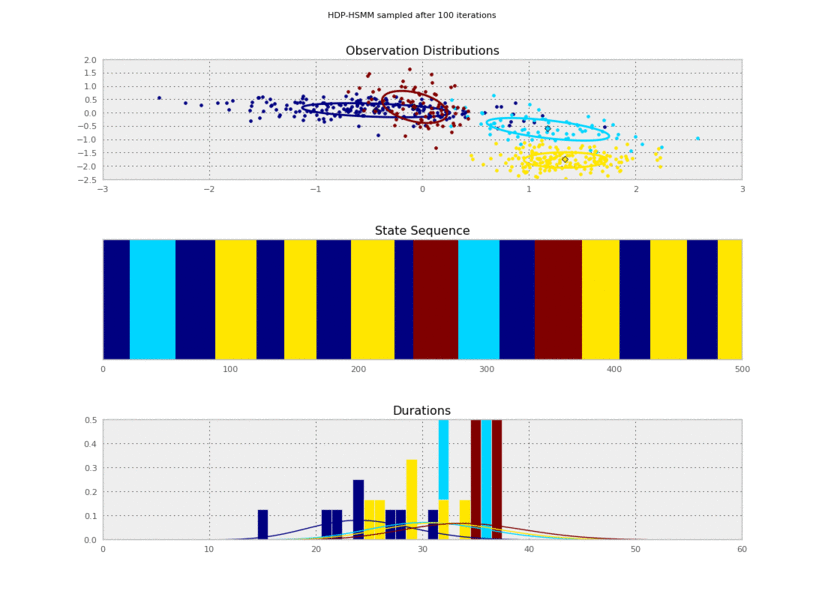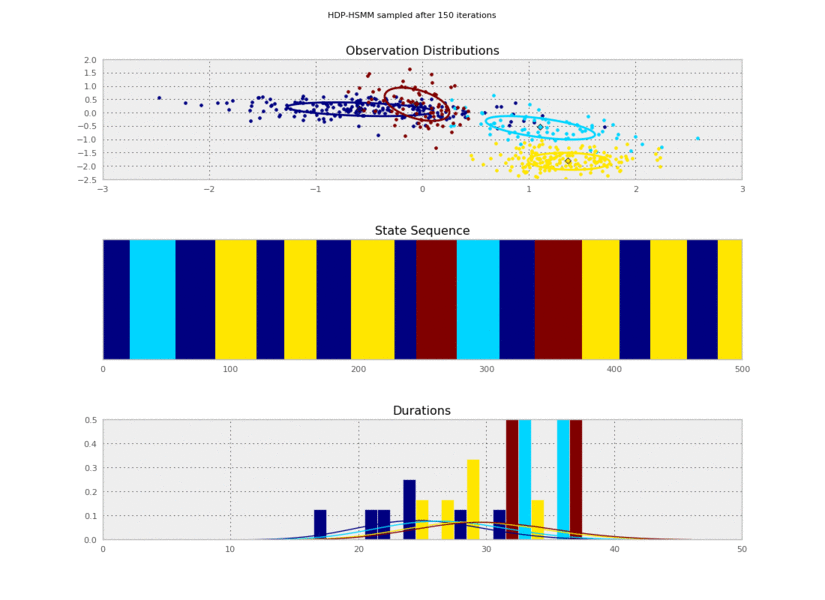
I generated these data from an HSMM that looked like this:
-
+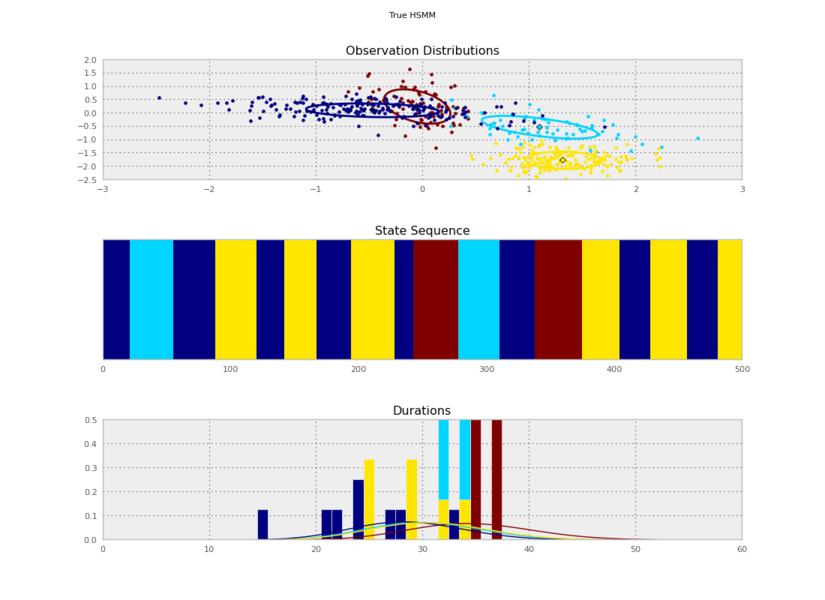
So the posterior samples look pretty good!