diff --git a/README.md b/README.md
index 1c05dde9e038b3dad339abddf0bb7479fcc0d3d9..baa0ebbdf27a1c567a3c0de60d509cf2e652f629 100644
--- a/README.md
+++ b/README.md
@@ -44,7 +44,7 @@ plt.plot(data[:,0],data[:,1],'kx')
plt.title('data')
```
-
+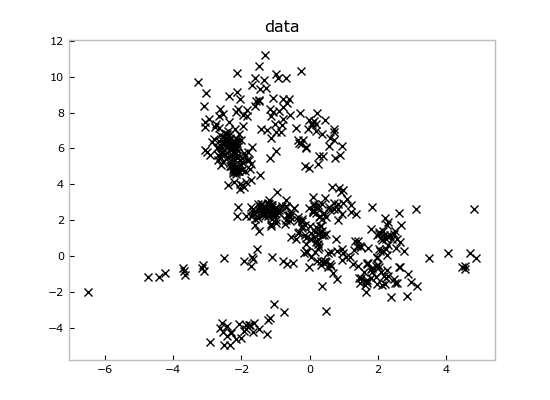
Imagine we loaded these data from some measurements file and we wanted to fit a
mixture model to it. We can create a new `Mixture` and run inference to get a
@@ -95,7 +95,7 @@ for scores in allscores:
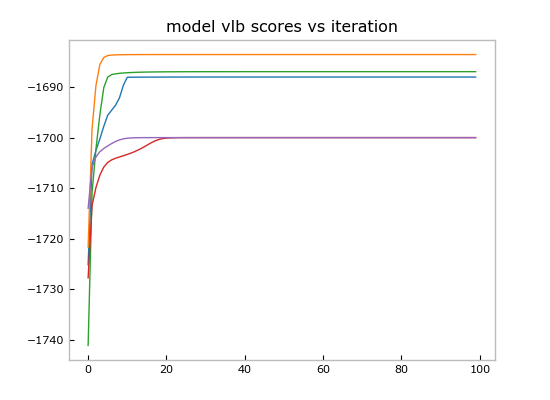
plt.title('model vlb scores vs iteration')
```
-
+
And show the point estimate of the best model by calling the convenient `Mixture.plot()`:
@@ -104,7 +104,7 @@ models_and_scores[0][0].plot()
plt.title('best model')
```
-
+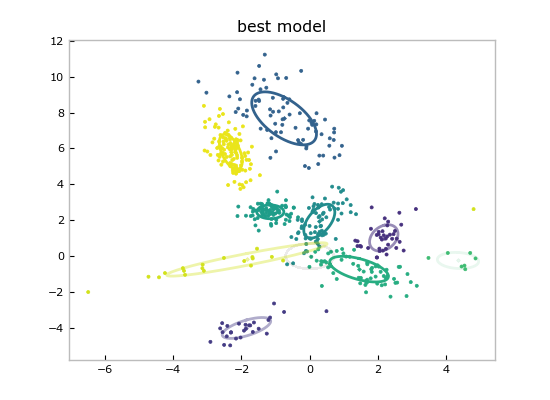
Since these are Bayesian methods, we have much more than just a point estimate
for plotting: we have fit entire distributions, so we can query any confidence